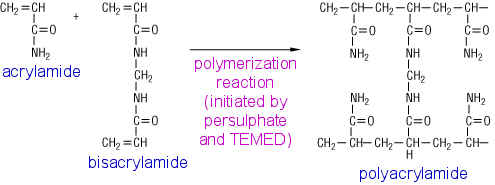
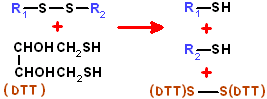Practice exercises
A) Influence of the applied voltage
Load the gel with all standards and any one of the unknown protein samples.
Choose in turn each of the voltages available in the simulator and run the electrophoresis until the tracking dye band (purple colour) approaches the lower end of the gel. Write down in each case the mobility of the standards.
1. Which is the effect of an increase in the voltage applied between both electrodes?
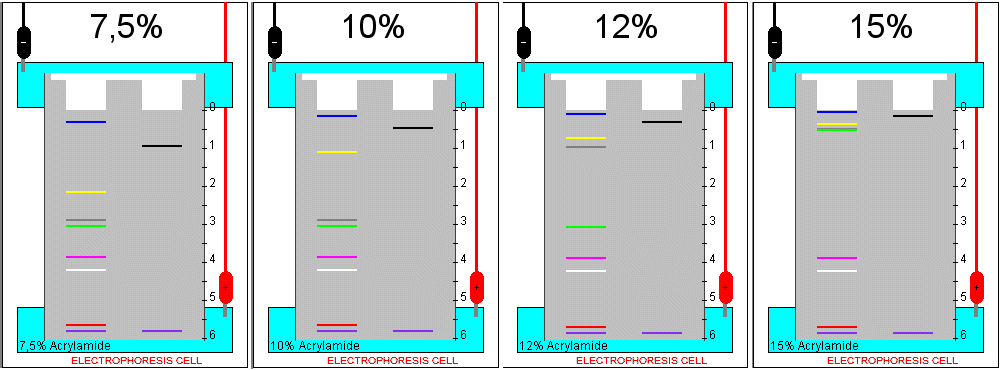
B) Influence of the acrylamide concentration
Load the gel with all standards and any one of the unknown protein samples.
Choose in turn each of the acrylamide concentrations available in the simulator and run the electrophoresis until the tracking dye band (purple colour) approaches the lower end of the gel. Write down in each case the mobility of the standards.
1. Which is the effect of an increase in the acrylamide concentration that forms the gel?
2. How can you interpret this result, in terms of the mechanism of separation?
C) Self-assessment of results for molecular mass
Choose one of the unknown protein samples, included in the simulator.
Load that sample in the gel, as well as all the standards.
Run the electrophoresis until the band of the tracking dye (purple colour) approaches the lower end of the gel.
Once finished, measure the mobility of the unknown protein and interpolate that value in the graph, to obtain the relative molecular mass.
To confirm that your results are correct:
| |
type here your result for the relative molecular mass |
| Unknown sample # 1 |
|
| Unknown sample # 2 |
|
| Unknown sample # 3 |
|
| Unknown sample # 4 |
|
| Unknown sample # 5 |
|
| Unknown sample # 6 |
|
| Unknown sample # 7 |
|
| Unknown sample # 8 |
|
| Unknown sample # 9 |
|
| Unknown sample # 10 |
|
D) Determination of quaternary structure
Load the gel in the simulator with all the standards and any one of the unknown protein samples (although you will not use the latter).
Run the electrophoresis until the band of the tracking dye (purple colour) approaches the lower end of the gel.
Once finished, access the graph of relative molecular mass against mobility and solve the following problems:
1. An enzyme is treated with SDS and beta-mercaptoethanol and analysed on a polyacrylamide gel with SDS; two bands are observed with relative mobilities of 0.47 and 0.59.
Thanks to some independent experiments involving size exclusion chromatography and ultracentrifugation, the molecular mass of this enzyme has been determined as 128 000.
Using the calibration line obtained in the simulator, estimate the molecular mass of both subunits of the enzyme and deduce which its quaternary structure is.
2. A protein treated with SDS but without beta-mercaptoethanol is analysed on a polyacrylamide gel with SDS, showing a single band with relative mobility of 0.21.
When the same protein is previously treated with SDS and beta-mercaptoethanol, electrophoresis displays two bands, with mobilities 0.48 and 0.61.
Using the calibration line obtained in the simulator, interpret the results.
3. (Advanced) If the protein is previously treated with trypsin, the same band is obtained in the absence of beta-mercaptoethanol, but four bands in its presence. Explain these results.
Angel Herráez. Part of the website Biomodel.uah.es
❖
Esta página está disponible también en español.

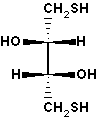 threo-2,3-dihydroxy-1,4-butanedithiol. Also known as Cleland's reagent.
threo-2,3-dihydroxy-1,4-butanedithiol. Also known as Cleland's reagent.