Practice exercise
Citrate synthase complexed with ligands
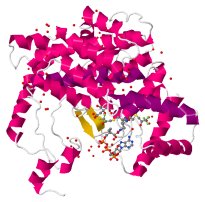 Load the PDB file with code 6CSC, but only one chain (monomeric enzyme):
Load the PDB file with code 6CSC, but only one chain (monomeric enzyme):
- sample file 6csc_monomer.pdb (right-click and choose download or save as)
- command in the console: load =6csc filter "*:A";
Using the mouse, find out the name of the ligands present; write those names down.
If it does not look like the image on the right, adjust the rendering:
- select all; spacefill 23%; wireframe 0.15;
(or popup menu )
- select protein; cartoon only;
(or popup menu then )
- select all;
Activate highlight for working more easy:
Now let's have a look at the citrate pocket:
In the console, type:
- select within(3.0, CIT); // see what got selected
- define ligandsite selected; // defining a "named atom set" allows to reuse later
-
menu
- maybe: display ligandsite; // hides everything else
then:
display all;
Try result setting other distances.
Better:
- select within(3.0, CIT) and not CIT; .
- define ligandsite selected;
How to measure distances:
- with the mouse: double-click on first atom, move to target atom, double-click to fix the measurement
- with a command: measure (atom1) (atom2); // learn how to specify atoms
- you can get a listing from the toolbar button
 Load the PDB file with code 6CSC, but only one chain (monomeric enzyme):
Load the PDB file with code 6CSC, but only one chain (monomeric enzyme): Load the PDB file with code 6CSC, but only one chain (monomeric enzyme):
Load the PDB file with code 6CSC, but only one chain (monomeric enzyme):