Both polypeptides, "A" or "M" and "B" or "H", are very similar in their structure, but have some differences in their sequence:
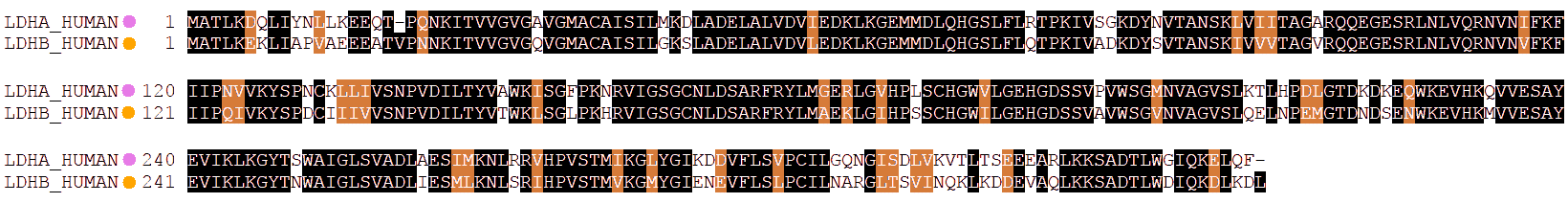
As a consequence, their physicochemical properties are a little different, among them the isoelectric point and the electric charge.
It may be noticed that the sequence of the A isoform has a higher number of residues with positive charge while B has more with negative charge. Therefore, B will have a more negative charge than A, and LDH tetramers will move faster towards the anode as they have more B subunits.
In other words, in order of decreasing mobility towards the anode: B4 > AB3 > A2B2 > A3B > A4
which is the same as H4 > MH3 > M2H2 > M3H > M4
or LDH-1 > LDH-2> LDH-3 > LDH-4
To ascertain this reasoning, and lacking an experimental measurement of the isoelectric point of each isoform, we may resort to computational prediction, based on known properties of each amino acid when they make part of a protein:
We hence check that B has a lower pI, is more negative than A for the same pH.
Angel Herráez. Part of the Biomodel.uah.es website
Sequences were obtained from UniProtKB; alignment was computed at T-Coffee and plotted using BoxShade (ExPASy).